3D Structure View
Loading...
More Info
GENERAL
TITLE
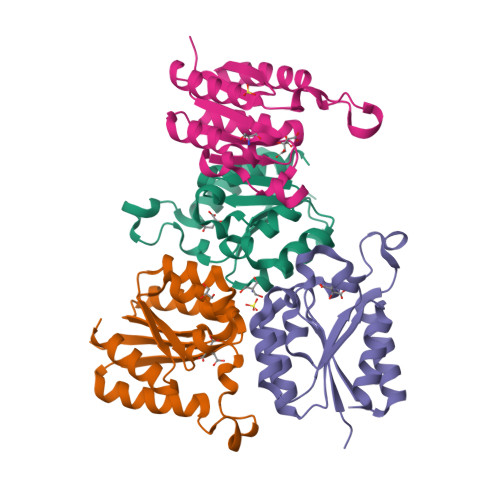
THE CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM ZYMOMONAS MOBILIS
DATE LAST MODIFIED
-
RELEASE
8 Jul 2020
OBSOLETE
CLASSIFICATION
BIOSYNTHETIC PROTEIN
EXPERIMENT TYPE
X-RAY DIFFRACTION
SPACE GROUP
H 3
RESOLUTION RANGE HIGH
2.34
RESOLUTION RANGE LOW
66.9
PUBMED ID
-
DOI
10.1039/C9NH00525K
PDBe
Chains
| Structure Chain | Target Protein | Gene | Expression System | Organism | Synonims | Actions |
|---|---|---|---|---|---|---|
| Q5NPJ9 | AROD, AROQ, ZMO0737, ZMO1_ZMO0737 | ESCHERICHIA COLI BL21(DE3) | ZYMOMONAS MOBILIS SUBSP. MOBILIS (STRAIN ATCC | 3-DEHYDROQUINASE,TYPE II DHQASE | ||
| Q5NPJ9 | AROD, AROQ, ZMO0737, ZMO1_ZMO0737 | ESCHERICHIA COLI BL21(DE3) | ZYMOMONAS MOBILIS SUBSP. MOBILIS (STRAIN ATCC | 3-DEHYDROQUINASE,TYPE II DHQASE | ||
| Q5NPJ9 | AROD, AROQ, ZMO0737, ZMO1_ZMO0737 | ESCHERICHIA COLI BL21(DE3) | ZYMOMONAS MOBILIS SUBSP. MOBILIS (STRAIN ATCC | 3-DEHYDROQUINASE,TYPE II DHQASE | ||
| Q5NPJ9 | AROD, AROQ, ZMO0737, ZMO1_ZMO0737 | ESCHERICHIA COLI BL21(DE3) | ZYMOMONAS MOBILIS SUBSP. MOBILIS (STRAIN ATCC | 3-DEHYDROQUINASE,TYPE II DHQASE |
NGL Viewer is used for the molecular visualization.
- AS Rose, AR Bradley, Y Valasatava, JM Duarte, A Prlić and PW Rose. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics: bty419, 2018. doi:10.1093/bioinformatics/bty419
- AS Rose and PW Hildebrand. NGL Viewer: a web application for molecular visualization. Nucl Acids Res (1 July 2015) 43 (W1): W576-W579 first published online April 29, 2015. doi:10.1093/nar/gkv402