3D structures for understanding mechanisms and designing drugs
Updated: 1/12/2021 - Updated numbers for structural snapshots and organisms. Updated images for CoV2, SARS, MERS and 3D structure.
3D structures are a powerful tool in understanding the molecular mechanisms of disease and the design and development of new drugs. canSAR-3D, the 3D structural component of canSAR, use artificial intelligence approaches to identify and predict the ‘ligandability’ (the propensity of a protein to bind a drug-like small molecule) of proteins with known 3D structure.
We map potential cavities and assess 3D-ligandability for all proteins released in the Protein Data Bank automatically update these in Coronavirus canSAR on a weekly basis. At the end of April 2020, canSAR contained >503,000 individual protein structural snapshots for >29,000 proteins from 2,893 organisms. For these, we applied our AI methods to assess >4 million cavities for ligandability. Using the same method, we also analysed the 3D structural interfaces of >117,000 biological complexes and 640,988 interface cavities. All results for all structural analyses and AI assessments are provided through a user-friendly interface on the canSAR website.
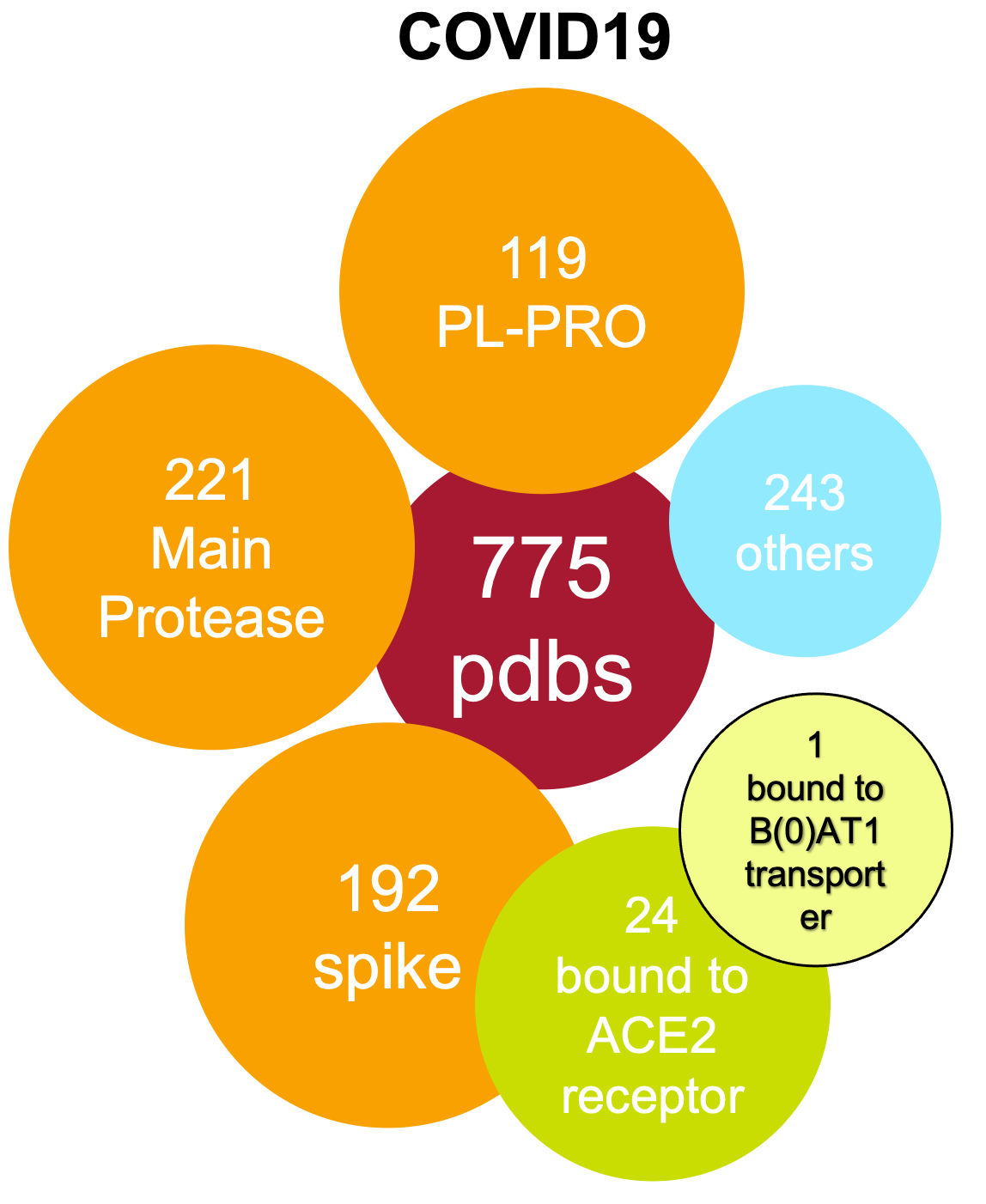 Covid-19
Covid-19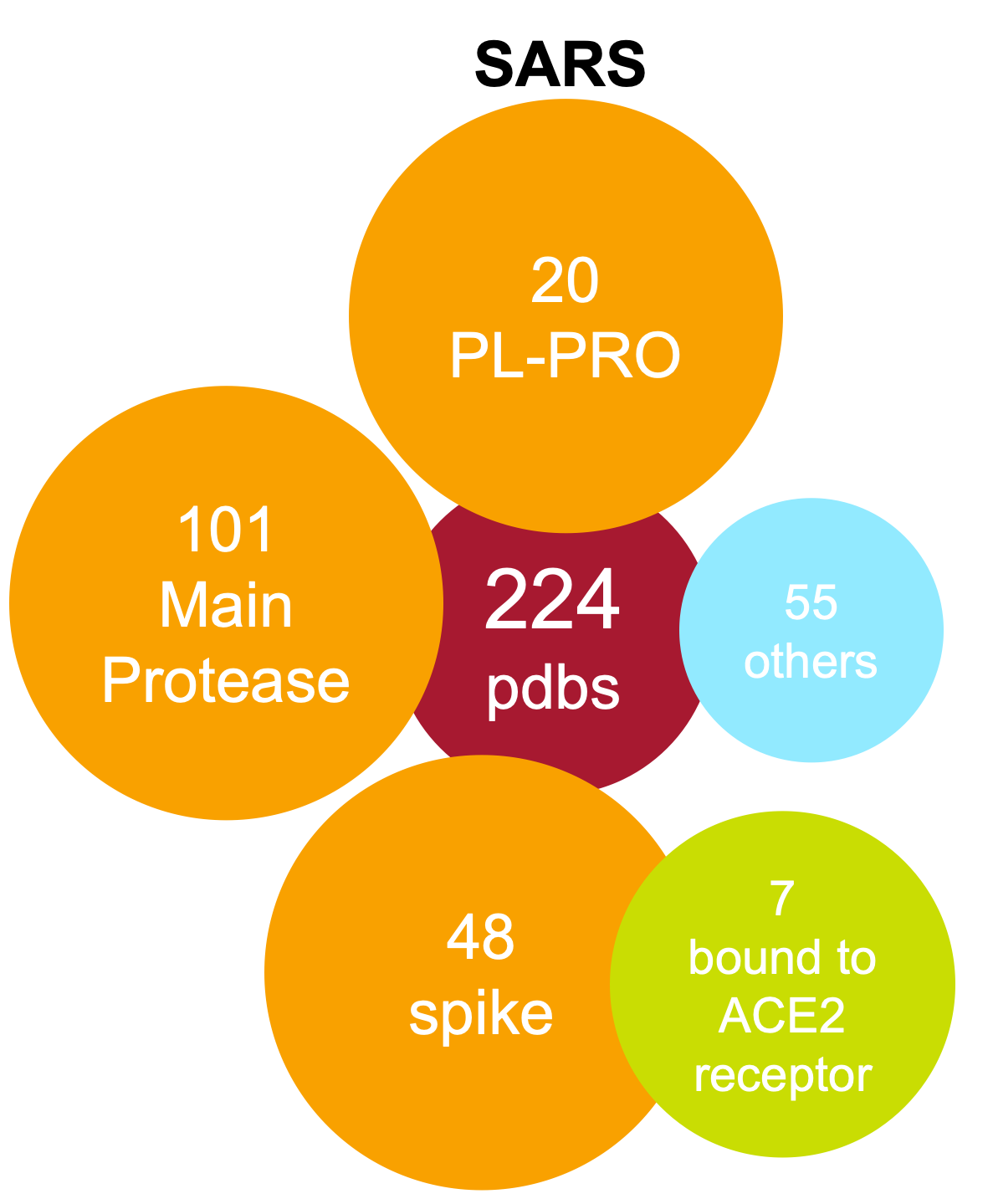 SARS
SARS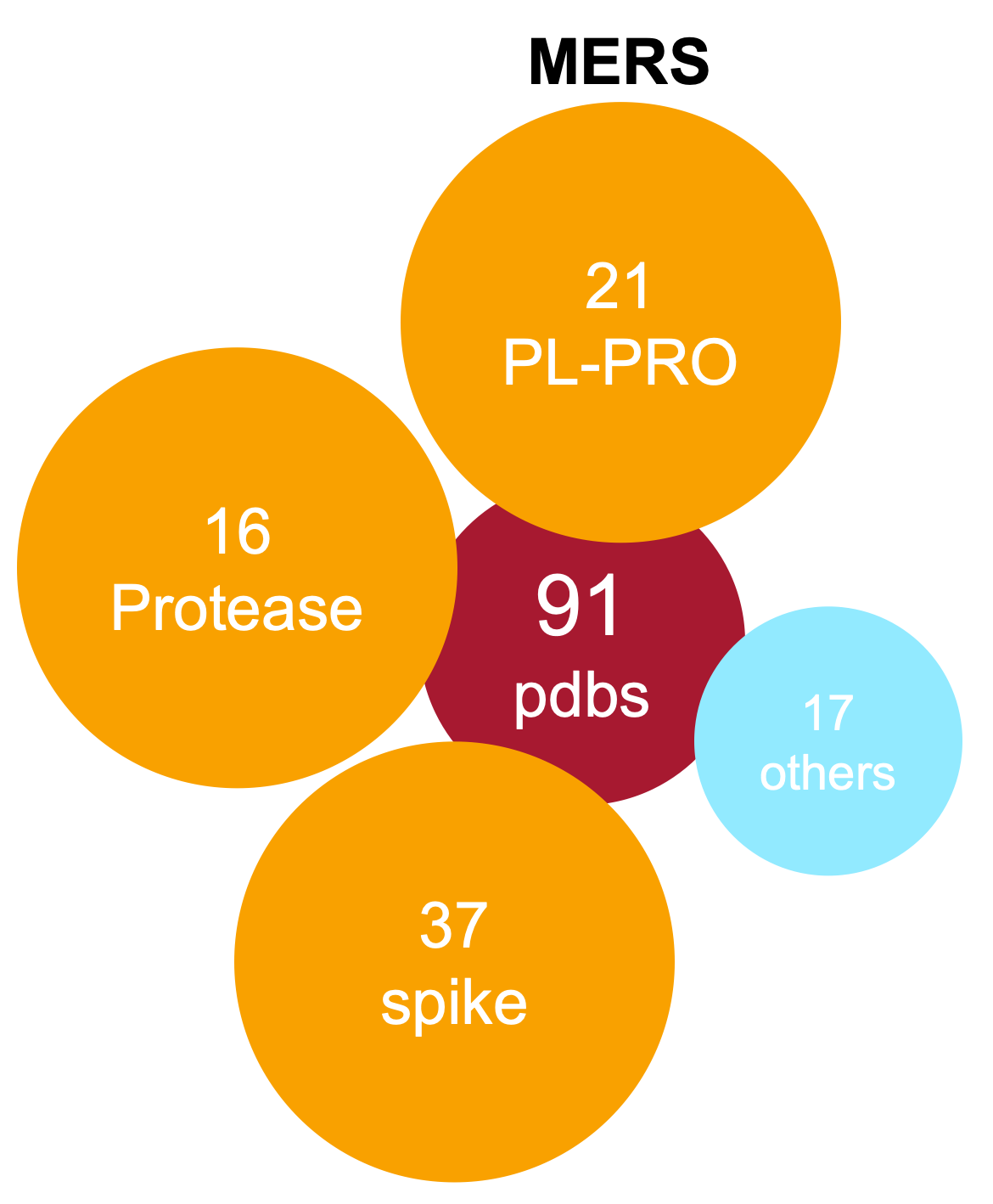 MERS
MERSAll 3D structures from Covid-19, SARS and MERS have been assessed) and a live, weekly updated summary of their ligandability analysis is reported in our structures report.
In particular, 229 COVID19, 443 SARS and 181 MERS protein chains have been analysed. 661 are from X-ray and 157 from cryo-EM and 35 from NMR. 80 human chains (belong to 6 different families) have been found to bind viral proteins.
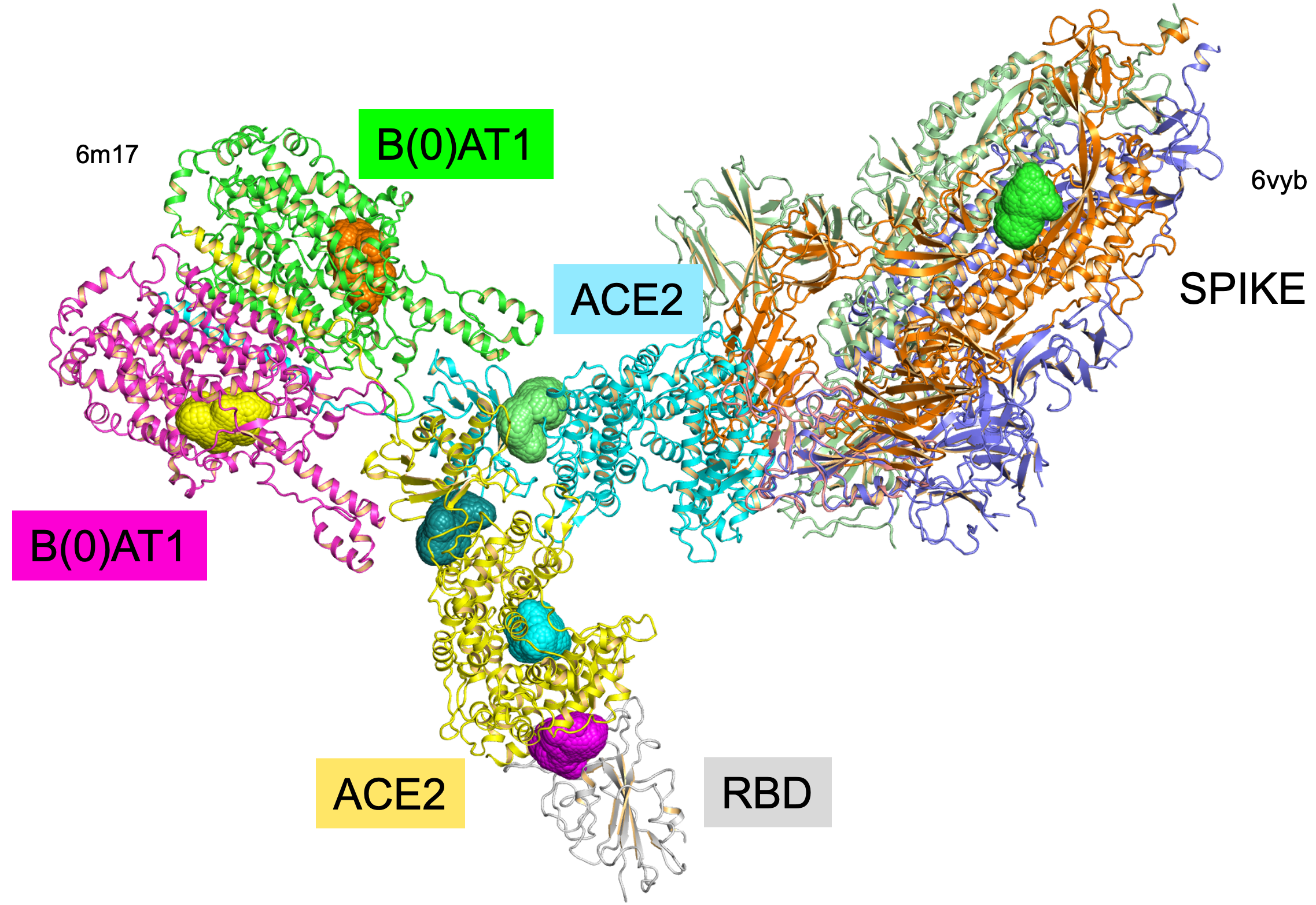 3d Structures related to coronavirus
3d Structures related to coronavirus
We analysed >8100 cavities on > 430 protein structures (857 PDB chains). As results of our analysis, we identify 284 ligandable pockets and 339 potential ligandable cavities at the interfaces of protein complexes (biological assemblies). We found novel ligandable cavities at the interface between the coronavirus Spike protein and their receptor ACE2, and at the complex’s interface with the Sodium-dependent neutral amino acid transporter B(0)AT1.
These weekly updated assessments are provided to support drug design and antibody discovery.